- Research News
Multiscale simulations unveil molecular mechanisms that shape brain plasticity
25 July 2022
Scientists of the Human Brain Project (HBP) have used simulation tools to uncover molecular mechanisms of a family of enzymes that is key to processes related to brain plasticity and learning. Their results, published in WIREs Computational Molecular Science, provide insights into the dynamics between so-called adenylyl cyclase enzymes and the proteins that regulate their activity.
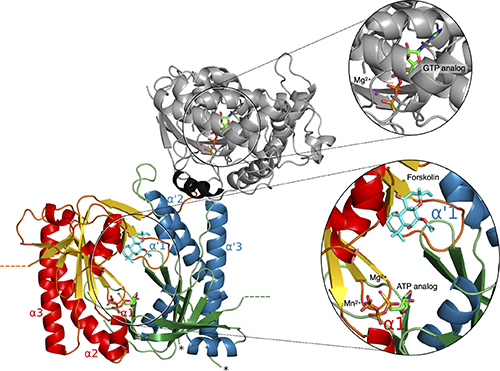
With their study, a collaboration between nine institutions from six countries, the authors present a proof of principle showing how using different molecular simulation techniques on a single enzyme and its regulators provides a detailed picture of its function – which would be difficult or impossible to obtain by empirical research.
Adenylyl cyclases (AC) constitute a family of enzymes that play key roles across all cells in the human body. Most importantly, AC catalyses the production of cyclic AMP, a messenger that regulates many fundamental cellular functions.
For the past few years, a team of Human Brain Project researchers from all over Europe has systematically investigated the structure, function, and dynamics of AC enzymes, focusing on a very important member of the family, the AC5 protein. They used a wide range of multiscale molecular simulation techniques, which enable the study of biological systems at the atomic level. Their findings were then incorporated in models of a network centred on AC5 that underlies memory reward processes in the brain. The authors uncover how AC5 interacts with its cellular partners, following binding of the neurotransmitters, dopamine (often associated with a rewarding stimulus) and acetylcholine, to their target receptors located on the cell surface.
They further show the ability of the AC5 enzyme to perform “coincidence detection” of the two neurotransmitters: a cyclic AMP response is produced only when the two neurotransmitters bind almost simultaneously to their receptors. This is a crucial requirement for synaptic potentiation, a process that is important for learning and memory.
For the future, the HBP scientists expect this type of research, that in this case required several years, to be carried out much more efficiently. Firstly, HBP scientists are developing molecular and subcellular simulation software infrastructures for the EBRAINS and FENIX services for multiscale brain simulation. Secondly, exascale computers, powerful computing systems that can perform one quintillion operations per second, are being installed all over the world. “We are working hard to adapt our biomolecular simulation codes towards this new architecture. These computers, if exploited properly by our codes, may expand tremendously the domain of applicability of biomolecular simulation,” says Paolo Carloni, head of the Institute for Computational Biomedicine at Forschungszentrum Jülich and co-author of the study. Europe’s first exascale computer will be in Forschungszentrum Jülich, the research centre where part of this team of researchers is based.
Link to the paper:
https://wires.onlinelibrary.wiley.com/doi/10.1002/wcms.1623
Text by Helen Mendes



