Macaque vision
Biologically meaningful simulation of early visual data processing in macaque cerebral cortex
This project investigates biologically meaningful models of macaque monkey visual cortical areas V1, V2 and V5. We aim at replicating receptive field properties using computational spiking neuron models. Visual environment is a rich sensory environment, macaque visual system is relatively well known, and with proper stimuli we can target specific neural subsystems. This creates a unique window for modeling cortical functions.
Project description
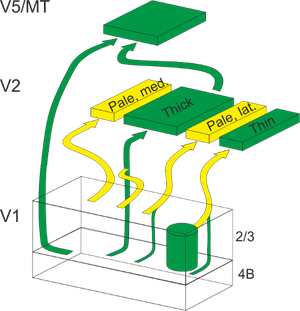
Our group simulates visual areas with anatomically, physiologically and biophysically meaningful (biomimetic) models. We have created the CxSystem simulation framework on top of the Brian2 simulator for quick development of models of cortical systems. Specifically, we aim to optimize our biomimetic model of the visual system to replicate known physiological receptive field properties of the macaque. Eventually, we aim to understand how biological tissue computes visual data, and how transformations in receptive field representations in single neurons are related to the computational tasks the system is facing. Our interest, compared to more abstract approaches, is in following the biological system. The abundant existing experimental data on the macaque visual system provides a map for our model development.
Objectives
We plan to build a functioning model of the cortical areas V1, V2, and V5 in the macaque monkey, using existing knowledge of thalamocortical and corticocortical connectivity. Our model will use comprehensive neuroinformatics from the macaque lateral geniculate nucleus and visual areas V1, V2 and V5. Our model structure will also be compared to Human Brain Project neuroinformatics data on the rat somatosensory cortex. The results of our simulations will be compared to existing physiological data from both macaque and human species.
Collaboration with HBP
Our main tool, the CxSystem, will become available to users of the Brain Simulation Platform. We have been using HBP neuroinformatics as well as eFel and NeuroM software tools for model construction.
Partnering Organisations


Biographies

Simo Vanni (Project Coordinator)
Dr. Vanni has studied visual cortex since 1995. After PhD from Helsinki University of Technology and University of Helsinki, he worked with Jean Bullier at Université Paul Sabatier in Toulouse as Marie Curie individual fellow. Earlier his work has included brain imaging, particularly fMRI. He was the scientific director of Aalto University Advanced Magnetic Imaging Centre 2010-2012. Since 2006 his work has included modeling and since 2012 simulations of visual cortex. He is currently affiliated with HUS Neurocenter, including clinical neurology at the Helsinki University Hospital, and to University of Helsinki.

Henri Hokkanen
MSc MD Hokkanen has a background in both mathematics and medicine. He currently has a graduate fellowship at the University of Helsinki, where he is working on his PhD project related to biomimetic models of cortical circuits.

Vafa Andalibi
Vafa Andalibi has programmed our main simulation tool CxSystem and the newer version CxSystem2. He has a background in computer engineering and received his double-major Master's degree in Biomedical Engineering and Communication Systems and Networks from Tampere University of Technology, Tampere, Finland. He is currently pursuing his PhD in Computer Science at Indiana University Bloomington, IN, USA, with a focus on security and privacy.



