DOPAMAP
Map of dopamine receptor positive cell types in the developing and adult mouse brain

Project Description
Dopamine is a neurotransmitter protein associated with motor control and motivational systems in the brain. The development of the dopaminergic system is yet poorly understood, but it is hypothesized to play an especially important role during childhood and adolescence, periods characterized by increased vulnerability to mental disorders. Changes in the dopaminergic system are particularly evident during this period. From its origin in the midbrain, the dopaminergic system branches out to reach large parts of the forebrain, heavily innervating regions of the prefrontal cortex and the striatum. Target cells in these areas express specific receptor types, making them sensitive to the dopaminergic input. A thorough understanding of the structural changes that occur in the dopaminergic system during normal development will be an important basis for studying disorders that affects it.
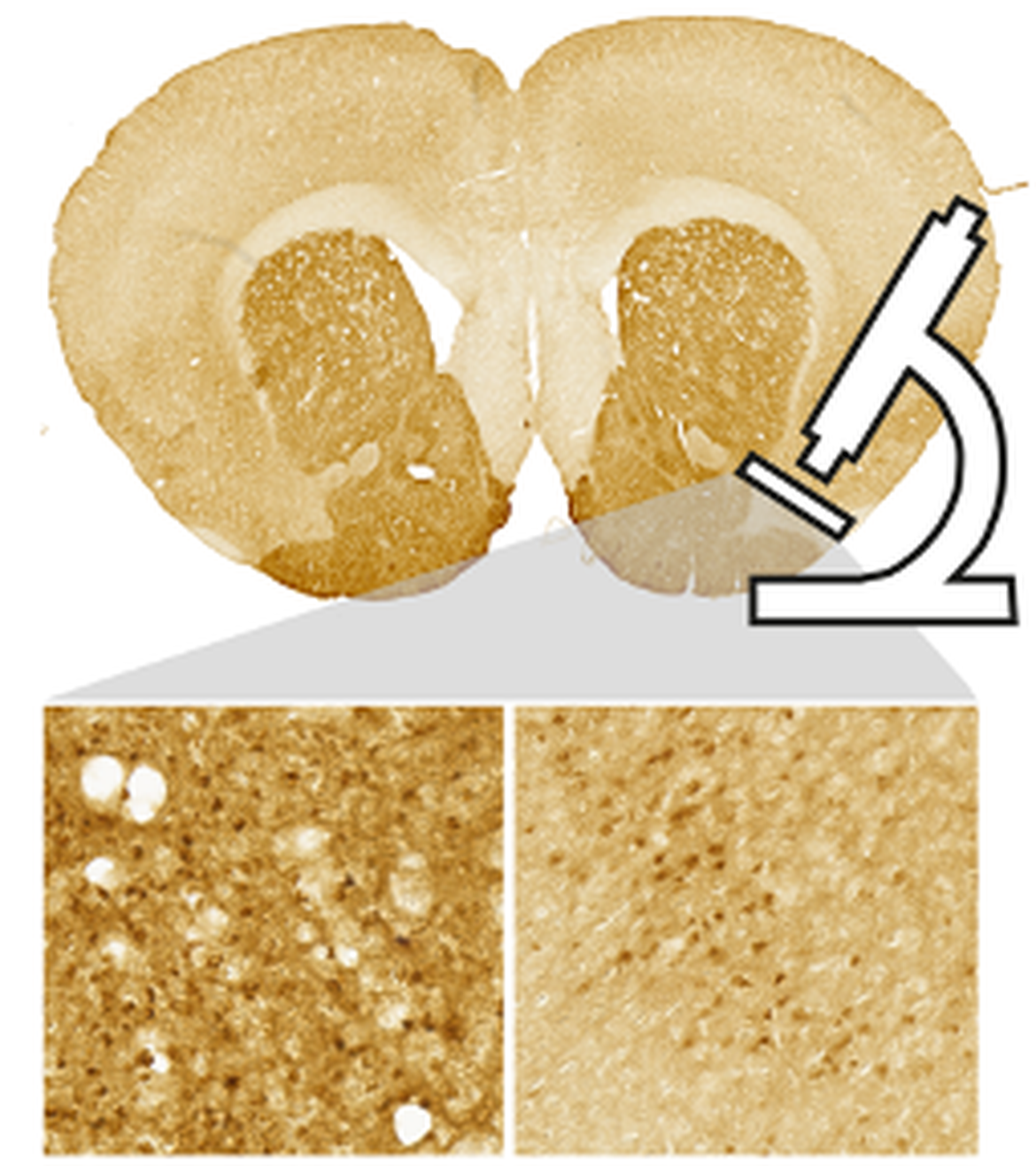
Headed by Associate Professor Jee Hyun Kim (Molecular Psychiatry Laboratory, Deakin University), DOPAMAP seeks to understand causes and effects of vulnerability to mental health conditions during adolescence. In this context, the lab has mapped cells positively marked with dopamine receptors 1 and 2 in developing and adult mice using immunohistochemistry and stereological quantification (Cullity et al. 2019). This is an extensive resource of >150 brains involving male and female mice aged 17, 25, 35, 49, and 70 days.
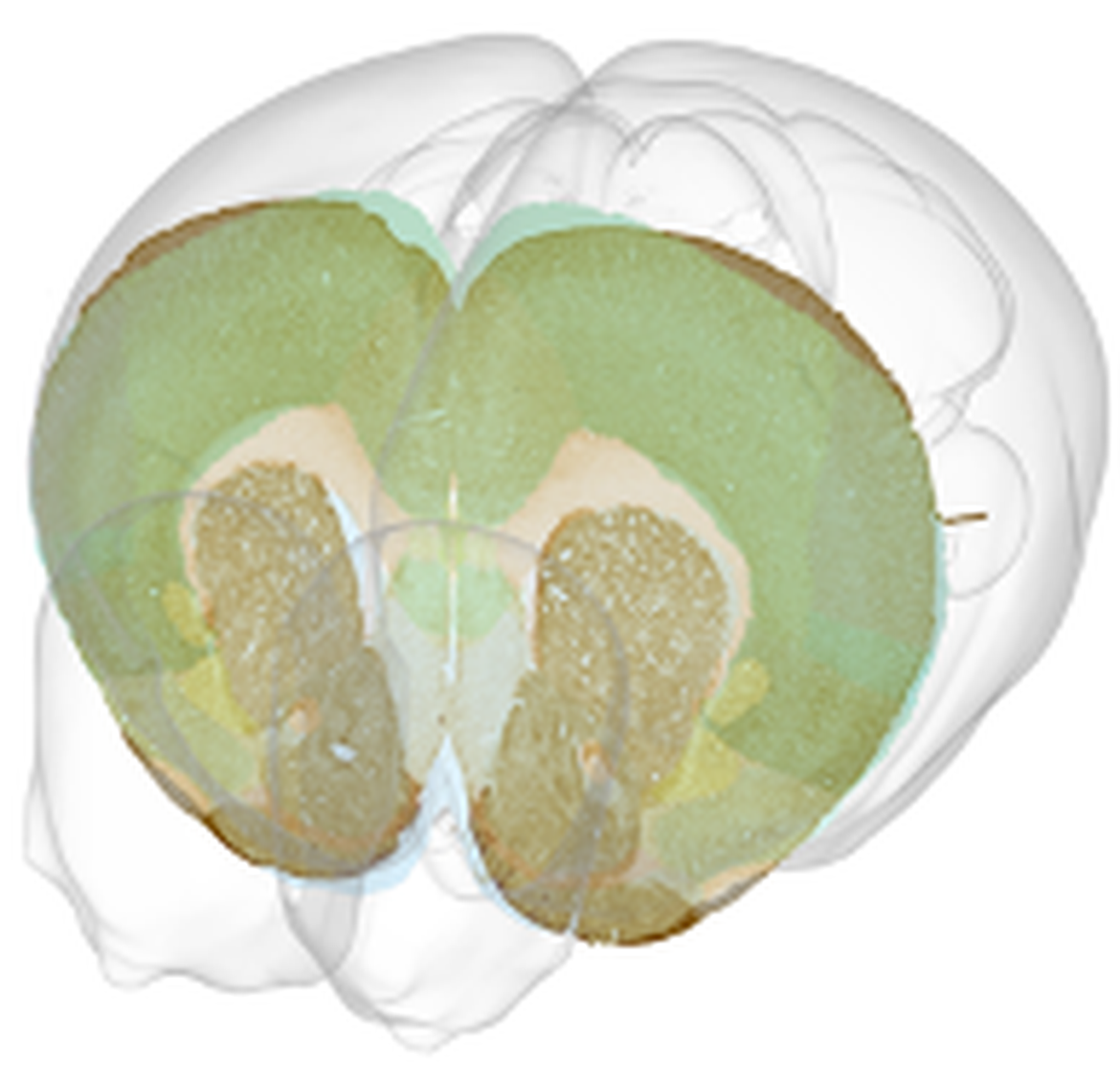
To allow the research community to re-use these data in new analyses, Associate Professor Kim and Associate Professor Leergaard (Neural Systems Laboratory, University of Oslo) have formed a new collaboration. Leergaard specializes in integration, management and sharing of neuroscientific data, with the aim of sharing processed tissue Cullity et al. (2019) in a Findable, Accessible, Interoperable and Re-usable way (Wilkinson et al., 2016). Therefore, the objective of DOPAMAP is to organise image data and associated metadata from these >150 animals. The data will be mapped to a common 3-D anatomical reference atlas (Wang et al., 2020), see example of a mapped section below (illustration made by use of the Scalable Brain Atlas composer (Bakker, Tiesinga, and Kotter 2015)) and make the data publicly available via the Neuroinformatics Platform of the HBP. Both male and female data are included and the subjects span five age groups, providing a uniquely rich dataset of the targeted cell types in the mouse brain. The key regions of the dopaminergic system covered by these data are also integral parts of the basal ganglia system. Thus, in addition to providing an important resource for researchers studying development and mental disorders, the data will also be useful for ongoing efforts to model the basal ganglia.
To ensure precise anatomical interpretation of the labelled cells, image registration tools (Puchades et al., 2019) are being adjusted to include atlas resources for developing brains. To this end, the project will make use of annotated reference atlas plates for the developing mouse brain, available from the Allen Institute, recently reconstructed and refined by Young and colleagues (2020). Validation of feature identification workflows are also in development.
This work will collect, organise, and preserve these data for future public data mining. The data will have strategic value for HBP ambitions to build a realistic model of the basal ganglia and implement this in a larger model of the whole brain.
References
Bakker R, Tiesinga P, and Kotter R. 2015. 'The Scalable Brain Atlas: Instant Web-Based Access to Public Brain Atlases and Related Content', Neuroinformatics, 13: 353-66.
Cullity ER, Madsen HB, Perry CJ, and Kim JH. 2019. 'Postnatal developmental trajectory of dopamine receptor 1 and 2 expression in cortical and striatal brain regions.', Journal of Comparative Neurology, 527: 1039-55.
Puchades M, Csucs G, Ledergerber D, Leergaard TB, and Bjaalie JG. 2019. 'Spatial registration of serial microscopic brain images to three-dimensional reference atlases with the QuickNII tool.', PLoS One, 14.
Wang Q, Ding S, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A et al. 2020. 'The Allen Mouse Brain Common Coordinate Framework: A 3D reference atlas'. Cell, 181: 1-18.
Wilkinson M, Dumontier M, Aalbersberg I, Appleton G, Axton M, Baak A, Blomberg N, Boiten J, da Silva Santos L, Bourne P et al. 2016. 'The FAIR Guiding Principles for scientific data management and stewardship.' Scientific Data, 3: 1-9.
Young D, Darbandi SF, Schwartz G, Bonzell Z, Yuruk D, Nojima M, Gole L, Rubenstein J, Yu W, and Sanders S. 2020. 'Automated Edge-Aware Refinement of Anatomical Atlases in 3D and Its Application to the Developing Mouse Brain.' bioRxiv: 2020.04.01.017665
Key facts
Time frame: 2020-2021
Origin: HBP Voucher Programme



